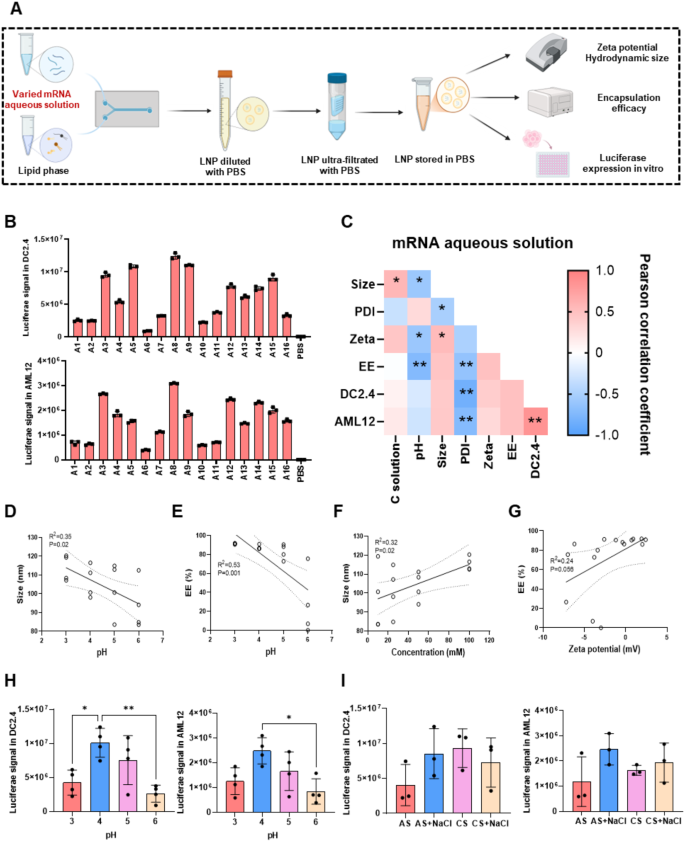
The effect of mRNA aqueous solution on LNP physicochemical properties and transfection in vitro
Firstly, we investigated the impact of pH (pH = 3, 4, 5, 6), salt type (citrate solution (CS), acetate solution (AS), CS plus NaCl, AS plus NaCl), and salt concentration (10, 25, 50, 100 mM) on the physicochemical properties and cellular expression of LNPs (Fig. 1A and B). The specific type of aqueous solutions was shown in Table S3. We measured the hydrodynamic diameter, PDI, zeta potential, and EE of LNPs prepared with different mRNA aqueous solutions (Table S3). The hydrodynamic diameter of these LNPs ranged from 84 to 120 nm, with a PDI between 0.08 and 0.21, zeta potential from − 7.2 to 2.4 mV, and EE ranging from 0 to 92%. The lowest EE was found in group A11 which the salt of mRNA aqueous solution was 50 mM CS and 130 mM NaCl with pH equaling 6. Moreover, the highest EE was achieved in group A13 with 91.8%, in which 100 mM CS with pH = 3 solution was used to disperse mRNA. Moreover, A13 also showed the highest size with 120.3 nm, while the smallest size was found in A1 (83.5 nm) which 10 mM CS (pH = 6) was used as mRNA aqueous solution. Then, we tried to figure out which factor impacted the physicochemical properties significantly and which factor or factors played the most crucial role. Therefore, we analyzed the correlation between the pH, salt concentration, and the physicochemical properties (Fig. 1C). The results showed a negative correlation between the pH of mRNA aqueous solution and the particle size, zeta potential and EE of the LNPs (Fig. 1C, D and E and Figure S1D).
Conversely, we only observed a positive correlation between the salt concentration of the mRNA aqueous solution and the particle size of LNPs (Fig. 1F). As the salt concentration increased, the particle size of LNPs slightly increased from 100 nm to 110 nm, likely due to the fusion between small vesicles under high ionic concentration [42]. This indicated that the concentration of salt had less significant effect compared with pH. Furthermore, we found that the type of salt showed no effect on the physicochemical properties (Figure S1H). Moreover, we found that EE and particle size increased with increasing zeta potential (Fig. 1G and Figure S1E). Additionally, Figure S1F and S1G demonstrated that EE and size were negatively correlated with PDI. These results suggested that the pH of mRNA aqueous solution was the most crucial role of mRNA aqueous solution affecting physicochemical properties which maybe because, as the pH increased, the degree of protonation LNPs decreased, leading to the exposure of mRNA on the LNP surface and resulting in the heterogeneous formation of mRNA/LNP complexes [35, 43, 44]. Consequently, EE and zeta potential decreased as the pH of the mRNA aqueous solution increased (Fig. 1C). Furthermore, high pH values contributed to the formation of empty liposomes, leading to smaller size and greater heterogeneity of LNPs [45] (Fig. 1C).
Subsequently, we examined the expression of these LNPs in AML12, DC2.4, and C2C12 cell lines (Fig. 1B, Figure S1A). The LNPs exhibited varying cellular transfection efficiency across different cell lines. Specifically, LNPs from groups A1, A2, A6, A10, and A11 showed relatively lower luciferase expression, while group A8 demonstrated the highest cellular transfection efficacy. Next, we investigated whether cellular expression correlated with the factors of the mRNA aqueous solution. As shown in Fig. 1H and Figure S1B, the pH of the mRNA aqueous solution significantly impacted LNP cellular expression. A decreasing trend in expression was observed in DC2.4, AML12, and C2C12 cells as the pH increased from 4 to 6. In DC2.4 cells, cellular expression was 2.9 times higher with a pH of 4 compared to pH = 6. In AML12 cells, the cellular expression of LNPs with mRNA aqueous solutions at pH = 5 and 6 decreased by 33% and 66% compared to pH = 4 (Fig. 1H). Similarly, in C2C12 cells, luciferase expression with pH = 4 mRNA aqueous solution was 3.9 times higher than that of pH = 6 (Figure S1B). Interestingly, LNPs with a pH of 3 did not follow this trend, possibly due to the instability of mRNA molecules at this low pH [46]. In contrast, the type and concentration of salts of the mRNA aqueous solution did not show any significant impact on cellular expression (Fig. 1I, Figure S1C).
Based on these findings, the pH of the mRNA aqueous solution emerged as the most critical factor influencing physicochemical properties and cellular expression of LNPs. Therefore, mRNA aqueous solution with pH = 4 was selected for further studies.
Influence of mRNA aqueous solution on mRNA/LNP. (A) Schematic of the LNP preparation method with different mRNA aqueous solutions. (B) Transfection efficiency of mRNA/LNP in DC2.4 cells and AML12 cells. (C) Correlation between mRNA aqueous solutions, physicochemical properties, and transfection ability. The correlation of pH and size (D), pH and EE (E), concentration of acid solution and size (F), Zeta potential and EE (G). (H) The impact of mRNA aqueous solution pH on the transfection of LNPs in DC2.4 cells and AML12 cells. (I) The effect of different acid type of mRNA aqueous solutions on the transfection of LNPs in DC2.4 cells and AML12 cells. Data were presented as mean ± SD. *p < 0.05, **p < 0.01
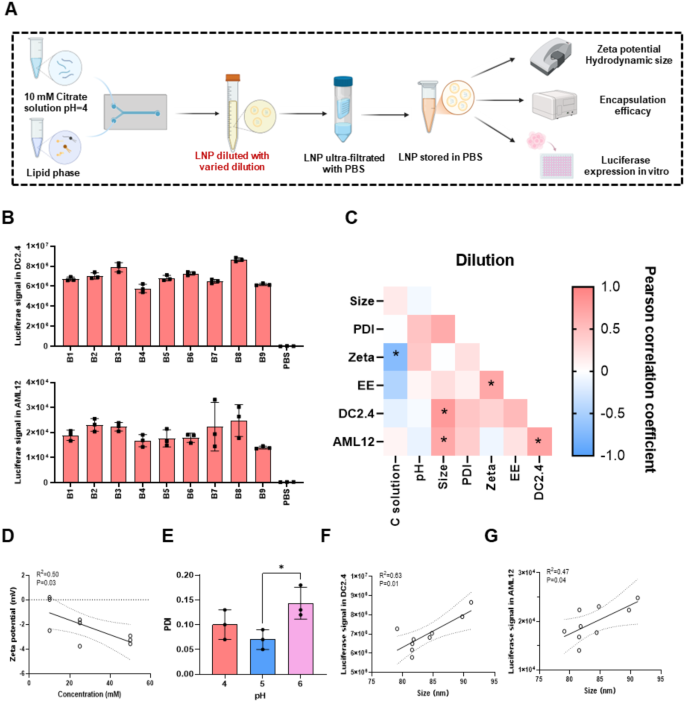
The effect of Dilution on physicochemical properties and transfection of LNPs
Subsequently, we examined the effect of pH (pH = 4, 5, 6), salt type (phosphate solution (PS), CS, AS), and salt concentration (10, 25, 50 mM) of the diluent on physicochemical properties and cellular expression of LNP (Fig. 2A). The dilutions were listed in Table S4. We tested the physicochemical properties of LNPs, luciferase expression at the cellular level (Table S4 and Fig. 2B), and explored the correlations among various indicators (Fig. 2C). Firstly, we evaluated the influence of dilutions on physicochemical properties. The size was approximately 84 nm and EE was around 91% (Table S4) with little difference between LNPs. Additionally, as shown in Fig. 2C and D, only the salt concentration of the diluent exhibited a negative correlation with the zeta potential of LNPs. pH, on the other hand, showed no significant correlation with the physicochemical properties, suggesting that the pH of the diluent was less important than the pH of the mRNA aqueous solution. Furthermore, LNPs formed at pH = 5 were more uniform, with a PDI of 0.07, compared to those formed at pH = 4 and pH = 6 (pH = 4: PDI = 0.10; pH = 6: PDI = 0.14) (Fig. 2E). The salt type did not significantly affect the physicochemical properties of LNPs (Figure S2A). Based on these results, we concluded that the dilution had a smaller effect on the physicochemical properties of LNPs compared to the mRNA aqueous solution.
In terms of cellular expression, the pH, salt type, and concentration of the diluent had no notable effects on LNP expression across different cell lines (Fig. 2C and Figure S2B-S2D), except in C2C12 cells, where the pH of the diluent influenced expression (Figure S2E). At pH = 4 (9.57*106), LNP expression in C2C12 cells was 20% or 24% higher compared to pH = 5 (7.75*106) or 6 (7.26*106), a phenomenon not observed in the other two cell lines. Moreover, we found that the expression of LNPs in AML12 and DC2.4 cell lines was positively correlated with particle size (Fig. 2F and G).
Based on these findings, there was no clear evidence indicating that any particular factor of dilutions had a significant impact on LNPs. Therefore, we proceeded to test all factors of the dilutions in combination with other types of solutions in further experiment.
The effect of dilutions on mRNA/LNP. (A) Schematic of the LNP preparation method with different dilutions. (B) Transfection efficiency of mRNA/LNP in DC2.4 and AML12 cells. (C) Correlation between physicochemical properties, transfection ability in vitro and dilutions of mRNA/LNP. (D) Correlation of dilution salt concentration and zeta potential. (E) The effect of dilution pH on PDI. The relationship of particle size and LNPs transfection in DC2.4 (F) and AML12 (G) cells. Data was presented as mean ± SD. *p < 0.05
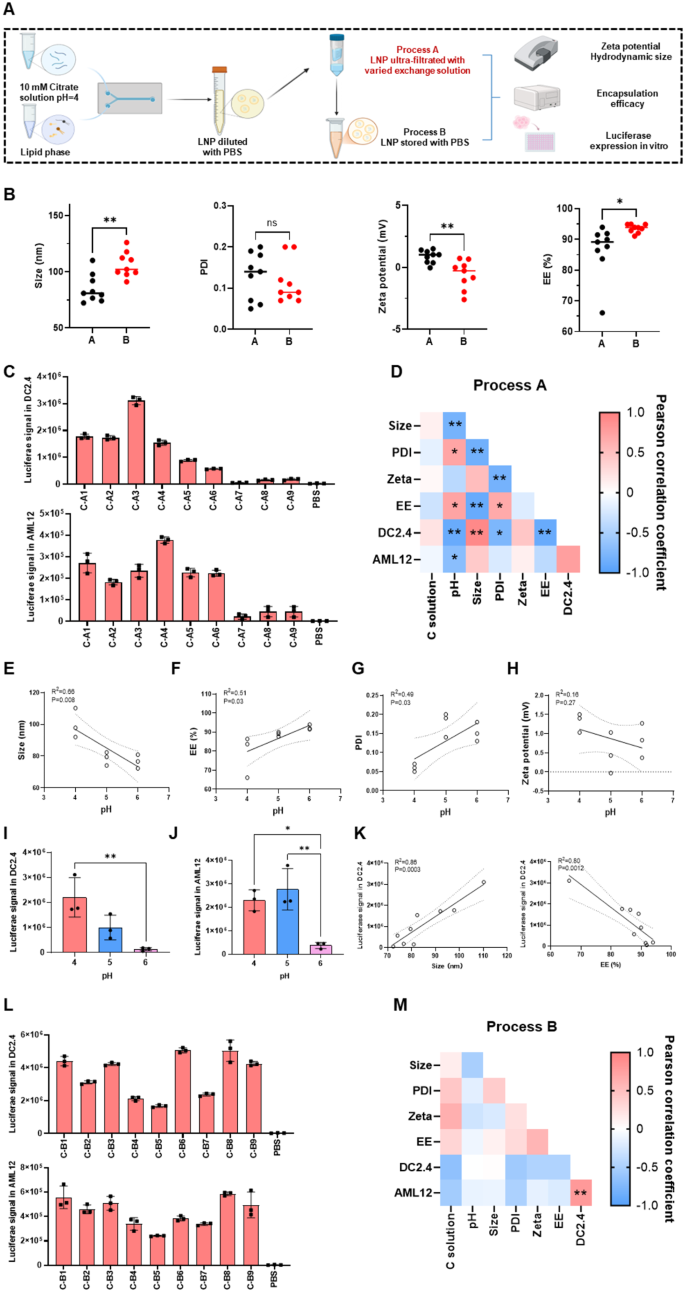
The effect of exchange solution on physicochemical properties and transfection of LNPs
We then studied the effect of different exchange solutions on the physicochemical properties and cellular expression of LNPs before (indicated as process A) and after (indicated as process B) exchange with the final storage solution (PBS) (Fig. 3A). The composition of exchange solutions used in each group were listed in Table S5.
Firstly, we compared the physicochemical properties of LNPs prepared by the two processes (Table S5). The data showed that LNPs prepared by process A had a notably smaller particle size (85 nm vs. 106 nm, p < 0.01) and EE (87% vs. 94%, p < 0.05) than those prepared by process B. However, process A LNPs exhibited a higher zeta potential (0.87 mV vs. -0.58 mV, p < 0.01). No significant difference in PDI was observed between the two methods (Fig. 3B). Next, we sought to determine which factors of the exchange solutions contributed to these differences. We assessed the influence of pH (pH levels of 4, 5, and 6), salt type (PS, CS and AS), and salt concentration (10, 25, and 50 mM) on particle size, zeta potential, and EE in both processes (Figure S3A). For particle size, LNPs prepared by process A with size below 100 nm, were significantly smaller than those prepared by process B, which was larger than 100 nm under all conditions. This may be due to the pH increasing from 4, 5, or 6 to 7 when adding the final storage solution-PBS. Regarding zeta potential, all LNPs in process A excepting those exchanged with 50 mM salt concentration or PS, showed significantly higher zeta potential than those in process B. This may be attributed to the variations of surface charge arising from different pH conditions. In terms of EE, LNPs formed by process A showed significantly lower EE than those formed by process B when the pH was 4 (79% vs. 94%, p < 0.0001), the salt concentration was 50 mM (83% vs. 94%, p < 0.001), or the salt type was PS (83% vs. 93%, p < 0.001). Under other conditions, no significant differences were observed.
We then examined how the pH, salt type, and salt concentration affected the physicochemical properties of LNPs in each process. The relationship between pH, salt type, concentration, and physicochemical properties in process A was analyzed (Fig. 3D and Figure S3B). We found that the pH of the exchange solutions was associated with all the tested physicochemical properties (Fig. 3D), except for zeta potential (R² = 0.16, p = 0.27, Fig. 3H). As shown in Fig. 3E, particle size exhibited a strong negative correlation with pH (R² = 0.66, p = 0.008). When the pH was 4, the particle size was around 100 nm, which decreased to approximately 80 nm at pH = 5 and 6. These results could be related to the different particle structures formed under various pH conditions, as reported previously [35]. Furthermore, EE decreased as the acidity of the exchange solution increased (Fig. 3F). At pH = 4, the EE was 14.8% lower compared to pH = 6. In addition, the PDI decreased from 0.15 to 0.05 as the pH dropped from 5 or 6 to 4 (Fig. 3G). In contrast, the type and concentration of salt in the exchange solutions had no significant impact on the physicochemical properties of LNPs (Fig. 3D and Figure S3B).
At the cellular expression level, pH was a critical factor. A negative correlation between pH and cellular expression was observed in DC2.4, AML12 and C2C12 cells (Fig. 3D, Figure S3D and S3E), with higher cellular expression seen when the pH was 4 or 5 (Fig. 3I and J and Figure S3E). Notably, DC2.4 cells were the most sensitive model among the three cell lines. Besides pH, the luciferase signal in DC2.4 cells was also significantly influenced by particle size and EE (Fig. 3D and K), which the luciferase expression in DC2.4 was elevated with the increasing of size (R2 = 0.86, p = 0.0003) and decreasing of EE (R2 = 0.8, p = 0.0012). Comparing with pH, other factors of exchange solutions such as salt type, concentration showed limited effect on the cellular expression of LNPs in both cells (Figure S3C).
In process B, there was no significant correlation between the exchange solutions and the physicochemical properties or cellular expression of LNPs (Fig. 3L and M). Except for C2C12, the luciferase signal was only influenced by salt type rather than pH and salt concentration (Figure S3G). The highest expression in C2C12 cells was observed when PS was used as the salt which was 50% higher than CS as the salt (Figure S3F and S3G). Moreover, the pH, salt type, and concentration showed limited effects on the expression of LNPs in the three cell lines (Figure S3F-I).
Based on these findings, exchange solution with 10 mM PS showed relatively higher protein expression and was selected as the preferred exchange solution for further studies.
The effect of exchange solutions on mRNA/LNP. (A) Schematic of the LNP preparation method with different exchange solutions. (B) The comparation of physicochemical properties of LNPs in process A and B. (C) Transfection efficiency of mRNA/LNP formed with process A in DC2.4 cells and AML12 cells. (D) Correlation of physicochemical properties, transfection ability and preparation parameters of mRNA/LNP in process (A) The influence of exchange solution pH on size (E), EE (F), PDI (G) and zeta potential (H) of LNPs formed with different exchange solutions. The effect of pH on luciferase expression in DC2.4 (I) and AML12 cells (J). (K) The relationship of LNP transfection in DC2.4 cells with size and EE. (L) Transfection efficiency of mRNA/LNP formed with process B in DC2.4 cells and AML12 cells. (M) Correlation of physicochemical properties, transfection ability and preparation parameters of mRNA/LNP in process (B) Data was presented as mean ± SD. *p < 0.05, **p < 0.01
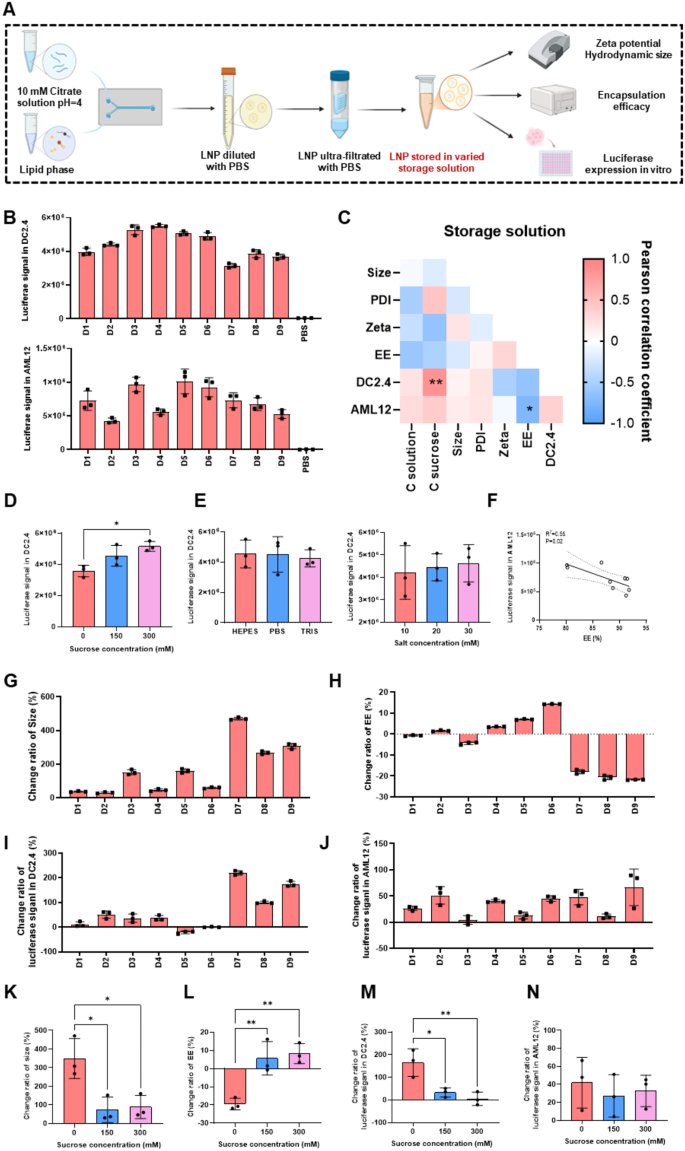
The effect of storage solutions on physicochemical properties and transfection of LNPs
Finally, we examined whether the physicochemical properties and cellular expression of LNPs were influenced by the storage solution (Fig. 4A). Specifically, we investigated the effect of salt type (Tris, HEPES, PBS), salt concentration (10, 20, 30 mM), and sucrose content (0, 150, 300 mM) of the storage solution (Table S6) on LNPs. The specific compositions and the physicochemical properties of each group were listed in Table S6. The data revealed that the storage solutions had no significant effect on the physicochemical properties of LNPs (Fig. 4C). The size of LNPs ranged from 79.5 to 99.6 nm, with near-neutral zeta potential, and EE ranged from 80.1 to 91.7% (Table S6).
Next, we evaluated the impact of the storage solutions on cellular expression. In DC2.4 cells, a positive correlation was observed between sucrose content and expression, with the highest LNP expression at 300 mM sucrose concentration, as shown in Fig. 4C and D. When the sucrose content increased from 0 to 300 mM, the luciferase signal increased by 31%. In contrast, the type and concentration of salt had no significant effect on expression (Fig. 4E). In AML12 cells, however, only the type of salt influenced expression, with lower cellular expression observed when HEPES was used compared to PBS and Tris (Figure S4A). Additionally, it was intriguing to note that EE was negatively correlated with cellular expression in AML12 cells (Fig. 4F). This could be due to relatively low EE promoting the release of mRNA from LNPs, thereby facilitating expression. Moreover, the type of storage solutions had no effect on the expression of LNPs in C2C12 cell line (Figure S4B).
mRNA-based drugs are unstable under vigorous shaking and long-term storage in solution form [47, 48], and as a result, mRNA drugs are typically preserved and transported after freezing or lyophilization. Additionally, under frozen condition, mRNA degradation occurs more slowly. However, the freeze-thaw process may disrupt the stability of mRNA/LNP formulations, leading to significant changes in their physicochemical properties and reduced expression both in vitro and in vivo [34]. Published studies had indicated that the storage solutions significantly impacted the freeze-thaw stability of mRNA/LNP drugs [40]. Therefore, we investigated the effect of different storage solutions on the stability of mRNA/LNP during the freeze-thaw process, providing a foundation for the storage condition of mRNA-based drugs (Fig. 4G-J).
We found that sucrose concentration significantly influenced both the changes in physicochemical properties and cellular expression of LNPs after the freeze-thaw process (Fig. 4K and N). Specifically, LNPs without sucrose protection tended to aggregate, forming larger particles and leaking mRNA, which decreased the EE after freeze-thaw (Fig. 4K and L). After freezing, LNPs in the 0% sucrose group experienced a 300% increase in size and a 20% decrease in EE. In contrast, when the sucrose concentration was increased to 150 mM or 300 mM, no significant changes in particle size or EE were observed (Fig. 4K and L).
Furthermore, sucrose concentration affected the expression of LNPs in DC2.4 cells after freeze-thaw, with the change in expression inversely proportional to the sucrose concentration (Fig. 4C and M). This phenomenon was not observed in AML12 cells, which could be related to cell-specific factors (Fig. 4N). Additionally, the type and concentration of salt in the storage solutions had no significant impact on particle size, EE, or cellular expression of LNPs before or after freeze-thaw (Figure S4C-F).
In this study, similar to previous reports [34], sucrose was identified as a critical factor influencing the stability of mRNA/LNPs. Although mRNA/LNPs without sucrose exhibited significant changes in size and EE, cell expression improvement was observed in DC2.4 cell lines. This could be attributed to the preference of DCs for phagocytosing the aggregates [49]. Based on these findings, a sucrose concentration of 300 mM was chosen for better freeze-thaw stability. Tris was selected as salt species, because it could provide additional protection for ionizable lipids [33] and showed no significant difference compared to other salts. The salt concentration was set to 20 mM to align with commercial formulations.
The effect of storage solutions on mRNA/LNP. (A) Schematic of the LNP preparation method with different storage solutions. (B) Transfection efficiency of mRNA/LNP in DC2.4 and AML12 cells. (C) Correlation between physicochemical properties, transfection ability in vitro and storage solutions of mRNA/LNP. The impact of sucrose concentration (D), salt type and concentration (E) of storage solutions on LNPs transfection in DC2.4 cells. (F) Correlation of EE with LNPs luciferase expression in AML12 cells. The change ratio of size (G), EE(H) and transfection of LNPs in DC2.4 (I) and AML12 (J) cells after freeze-thaw. The effect of sucrose concentration of storage solutions on the change ratio of size (K), EE (L), luciferase expression in DC2.4 (M) and AML12 cells (N). Data was presented as mean ± SD. *p < 0.05, **p < 0.01
The impact of integrated Preparation solutions on physicochemical properties and transfection of LNPs
After systematically evaluating the individual effects of various preparation solutions on the physicochemical properties and transfection efficiency of mRNA/LNPs, we observed several intriguing phenomena. However, these findings were derived under conditions where other preparation solutions remained constant, raising the question of whether these results would persist when multiple preparation parameters were simultaneously varied. Additionally, certain results from earlier experiments exhibited limitations in parameter ranges, which could compromise the robustness of the conclusions. To address these uncertainties, we integrated the optimized preparation conditions based on the performance of physicochemical properties and in vitro transfection efficiency, and subsequently redesigned the preparation process (Fig. 5A, Table S7). Given the demonstrated benefits of pH 4 mRNA solutions, we selected a range of pH 4 mRNA solutions, varying in salt concentrations (10, 25, 50, 100 mM) and salt types (CS, CS + NaCl, AS, AS + NaCl), as candidates for further evaluation. Although no significant differences were observed in the dilution solution, we expanded the screening scope by incorporating diverse salt species (PS, CS, AS, Tris), salt concentrations (10, 25, 50 mM), and pH levels (4, 5, 6, 7) to ensure comprehensive coverage.
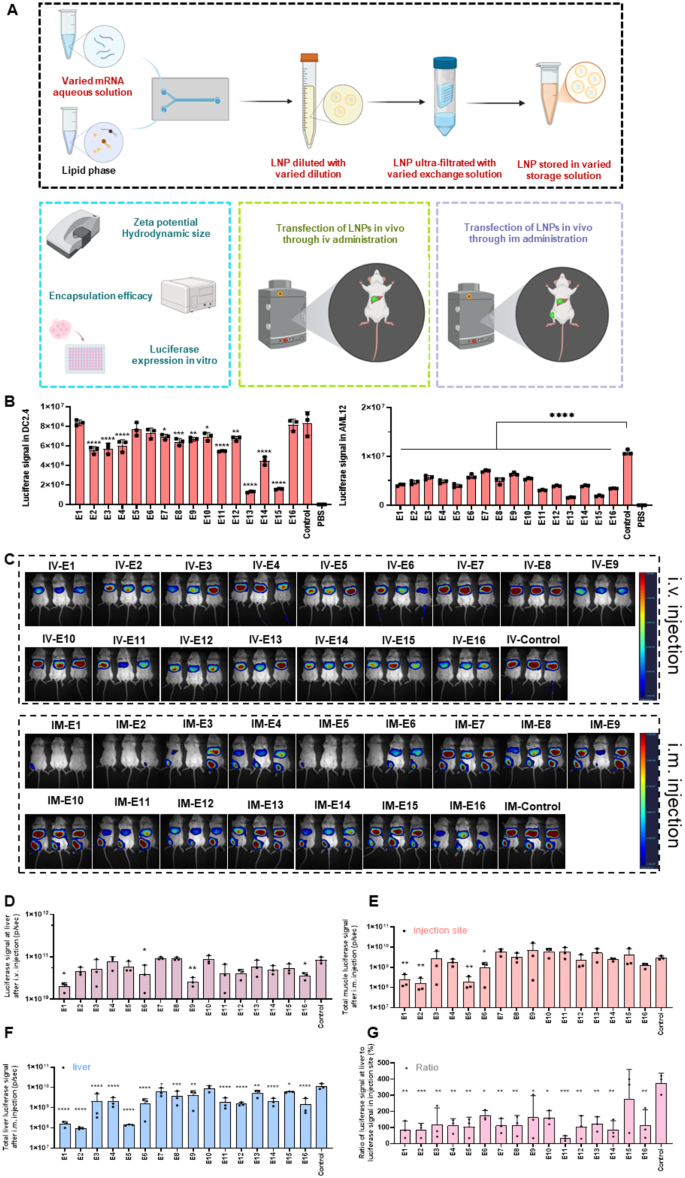
Furthermore, based on preliminary findings indicating that the pH of the exchange solution significantly influenced mRNA/LNP formation, we narrowed the screening parameters to confirm this observation by designing exchange solutions with pH 4, 5, 6, and 7 PS. Lastly, while 300 mM sucrose was identified as a critical component for stabilizing mRNA/LNPs during freeze-thaw cycle, the potential interaction between sucrose and other excipients in the storage solution remained unexplored. To address this, we combined 300 mM sucrose with Tris solutions of varying pH levels to further assess the influence of storage solution composition on mRNA/LNP stability and performance. As a control, we employed commonly used preparation conditions, where 10 mM CS (pH 4) served as the mRNA aqueous solution, and PBS was used for dilution, exchange, and storage. We investigated the physicochemical properties, in vitro transfection efficiency (Fig. 5B and Figure S5A), and in vivo expression (Fig. 5C, D and G) of LNPs formed by different solutions and analyzed their correlation (Fig. 6A).
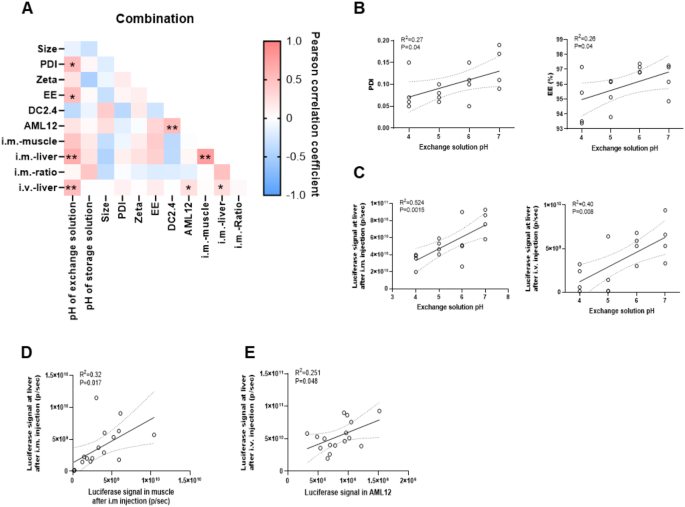
In terms of physicochemical properties, the hydrodynamic diameter of these LNPs ranged from 93.0 to 122.6 nm, with a PDI between 0.05 and 0.19, zeta potential ranging from 0.4 to 2.7 mV, and EE ranging from 93.5 to 97.4%, which were higher than those of the control group (EE = 88.2%, Table S7). Then we analyzed the correlation between physicochemical properties of LNPs and factors of solutions. Figure 6A and B showed that the pH of the exchange solution was the most critical factor, showing a positive relationship with PDI and EE (p = 0.04). However, the type and concentration of salt in the mRNA aqueous solution, as well as the addition of sodium chloride, had no effect on the particle size, PDI, or EE of LNPs (Figure S5B), except when the salt type was CS, in which case the EE decreased by 6% compared to when AS was used. The salt concentration in the diluent, exchange solution and storage solution had no impact on the physicochemical properties of LNPs (data not showed).
Regarding cellular expression, the control group demonstrated relatively better performance in all cell lines, particularly in AML12 and C2C12, in which the control group exhibited the highest luciferase expression (Fig. 5B and Figure S5A). In DC2.4 cells, only E1, E5, E6, and E16 showed comparable luciferase expression to the control group (Fig. 5B). However, we did not find any obviously correlation between factors of solutions and cellular expression as showed in Fig. 6A.
Subsequently, we investigated the expression of different LNPs in mice after i.v. or i.m. injection. We found that, with the same lipid formulation, altering the solutions used during the LNP preparation process could significantly impact the expression intensity in mice (Fig. 5C and D-G). After i.v. injection, E4, E7, E8, and E10 exhibited comparable expression to the control group, while E1, E6, E9, and E16 groups demonstrated significantly decreased luciferase signals (Fig. 5D).
Regarding i.m. administration, the results of luciferase expression differed considerably from those observed with i.v. administration. For luciferase signal at the injection site, groups E1, E2, E5, and E6 exhibited significantly lower expression compared to the control group (Fig. 5E). In terms of luciferase expression at liver after i.m. injection, all groups except E10 showed significantly lower luciferase expression than the control group (Fig. 5F). Consequently, most experimental groups, except E15, had significantly lower biodistribution outside the injection site after i.m. administration (Fig. 5G). These findings indicated that the solutions used to form LNPs not only affected their physicochemical properties and in vitro transfection efficiency but also influenced protein translation in vivo.
Next, we analyzed which factors of the preparation solutions could affect the expression of LNPs in vivo. The data showed that increasing the pH of the exchange solution enhanced mRNA expression at liver, regardless of whether the administration through i.v. (p = 0.008) or i.m. (p = 0.0015) (Fig. 6C). However, the mRNA aqueous solution, dilution, exchange solution and storage solution had no significant effect on in vivo expression (Figure S5C-E). Moreover, there was a strong correlation between protein expression at the injection site and that at liver, with a p-value of 0.017 after i.m. injection (Fig. 6D).
Additionally, we found that the expression of LNPs in AML12 cells in vitro was highly consistent with that observed at liver after i.v. administration in vivo (p = 0.048, Fig. 6E), suggesting that AML12 cells could be used to predict LNP expression in vivo.
The effect of different combined solutions on mRNA/LNP. (A) Schematic of the LNP preparation method with different integrated solutions. (B) Transfection efficiency of mRNA/LNP in DC2.4 and AML12 cells. (C) Bioluminescent images and quantification of mice 6 h after i.v. (upper panel) and i.m. injection (lower panel) with Luc mRNA/LNP (5 µg mRNA per mouse) by the IVIS imaging system. (D) Quantitively analyzing the luciferase signal in mice after i.v. injection. Quantitively analyzing the luciferase signal at injection site (E), liver (F) and the ratio between liver and injection site (G) after i.m. injection. Data was presented as mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001
The stability of LNPs formed with different solutions after freeze-thaw
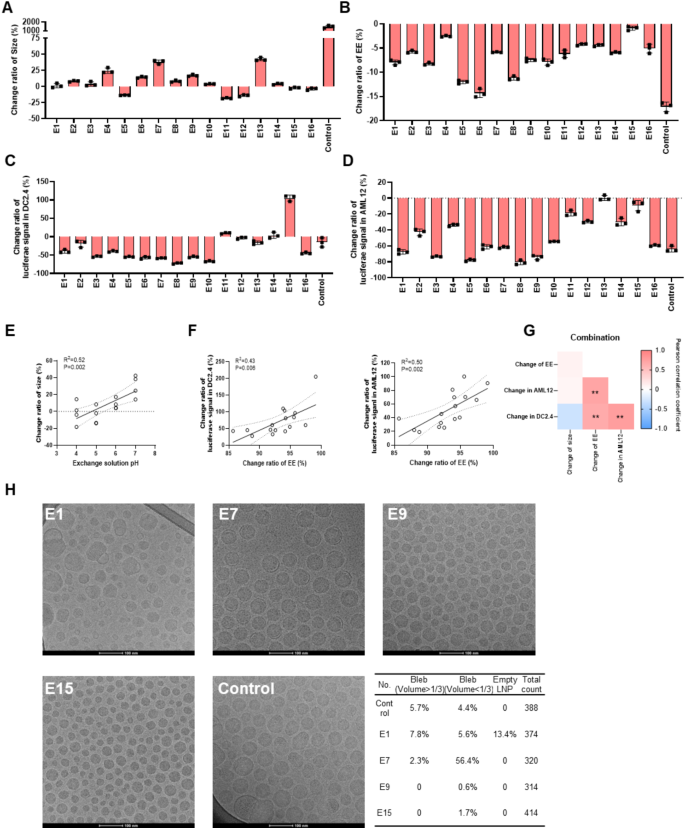
Subsequently, the stability of LNPs prepared using different preparation solutions was evaluated under freeze-thaw stress. We found that the size of the control group, which lacked sucrose protection, increased more than 10 times after freeze-thaw (Fig. 7A). In contrast, the size of some LNPs dispersed in a Tris/sucrose solution changed by less than 25% compared to the initial size, with the exception of E4, E7, and E13 (Fig. 7A), which could be related to the pH of the exchange solution (Fig. 7E). Additionally, most of the changes of EE were below 10%, except for E5 (12.0%), E6 (14.3%), E8 (11.2%), and the control group (17.6%) (Fig. 7B).
Next, expression changes in vitro were evaluated to assess the resistance of LNPs to the freeze-thaw process. To our surprise, although the physicochemical changes of most LNPs met the acceptable criteria, more than 25% of transfection reduction was observed in both DC2.4 and AML12 cells, except for E11, E13, and E15 (Fig. 7C and D). Furthermore, we found that the change ratio of expression in AML12 and DC2.4 cells was positively correlated with the change ratio of EE (Fig. 7F and G). Among the different LNP formulations, LNP E15 exhibited the best stability.
We then sought to determine why LNPs with the same lipid composition but different preparation solutions exhibited varying physicochemical properties and expression profiles in vitro and in vivo. We hypothesized that different preparation solutions may impact the structure of LNPs, subsequently influencing their performance. To test this hypothesis, samples from E1, E7, E9, E15, and the control group — chosen for their distinct performance — were further characterized using cryo-TEM. As shown in Fig. 7H, the structure of LNPs from different preparation conditions varied significantly. For example, particles from the control group exhibited nearly 10% of bleb structures (5.7% of particles with blebs occupying more than one-third of the LNP volume, and 4.4% of particles with blebs occupying less than one-third of the LNP volume). In addition to the bleb structure, a notable number of empty LNPs smaller than 30 nm were observed in the E1 group. Furthermore, a higher proportion (> 50%) of LNPs carrying blebs was observed in the E7 group compared to other groups. Although particles from the E9 and E15 groups exhibited a more homogeneous structure than those from E1, E7, and the control group, the shape and internal features of the LNPs from E1 and E7 require further investigation to understand the differences. The structural differences between the LNPs may partially explain the varied performance of LNPs prepared under different conditions, but additional experiments are needed to verify this hypothesis.
The relationship of factors in integrated solutions with physicochemical properties and transfection of LNPs. (A) Correlation between physicochemical properties, transfection in vitro, in vivo and factors of solutions. (B) The relationship between pH of exchange solutions with PDI and EE. (C) Correlation between pH of exchange solutions and luciferase expression at liver by i.v. or i.m. administration. (D) The relationship between luciferase expression at muscle and at liver through i.m. injection. (E) Correlation between transfection of LNPs in AML12 cells and at liver by i.v. injection. Data was presented as mean ± SD. *p < 0.05, **p < 0.01
The stability of LNPs formed with different solutions after freeze-thaw. (A) The size change ratio of LNPs after freeze-thaw. (B) The EE change ratio of LNPs after storing at -20℃ and thawing. The change ratio of luciferase expression in DC2.4 (C) and AML12 cells (D). (E) The relationship of exchange solution pH and size change ratio of LNPs. (F) Correlation of EE change ratio and the luciferase expression change ratio of LNPs after freeze-thaw in DC2.4 and AML12 cells. (G) Correlation between the change ratio of physicochemical properties and transfection in vitro. (H) The morphology of LNPs in different groups by cryo-TEM. Data was presented as mean ± SD. **p < 0.01